Advanced community ecological data analysis using vegan
Gavin L. Simpson
July 9th, 2020
Welcome
Logistics
Slides
Slide Deck: bit.ly/adv-vegan Sources: github.com/gavinsimpson/advanced-vegan-webinar-july-2020
Direct download a ZIP of everything: bit.ly/adv-vegan-zip
Unpack the zip & remember where you put it
Q & A
Add questions to Google Doc: bit.ly/adv-vegan-qa
Recording
Livestream will be recorded — link will be emailed to you later today
Today's topics
- Constrained ordination
- Canonical Correspondence Analysis
- Redundancy Analysis
- Partial constrained ordination
- Model Building
- Model selection
- Permutation tests
- Restricted permutation tests
- PERMANOVA
- Diagnostics
Constrained Ordination
CCA
Canonical Correspondence Analysis
CCA is the constrained form of CA; fitted using cca()
Two interfaces for specifying models
- basic;
cca1 <- cca(X = varespec, Y = varechem) - formula;
cca1 <- cca(varespec ~ ., data = varechem)
RDA is the constrained form of PCA; fitted using rda()
Formula interface is the more powerful — recommended
Canonical Correspondence Analysis
cca1 <- cca(varespec ~ ., data = varechem)cca1## Call: cca(formula = varespec ~ N + P + K + Ca + Mg + S + Al + Fe + Mn +## Zn + Mo + Baresoil + Humdepth + pH, data = varechem)## ## Inertia Proportion Rank## Total 2.0832 1.0000 ## Constrained 1.4415 0.6920 14## Unconstrained 0.6417 0.3080 9## Inertia is scaled Chi-square ## ## Eigenvalues for constrained axes:## CCA1 CCA2 CCA3 CCA4 CCA5 CCA6 CCA7 CCA8 CCA9 CCA10 CCA11 ## 0.4389 0.2918 0.1628 0.1421 0.1180 0.0890 0.0703 0.0584 0.0311 0.0133 0.0084 ## CCA12 CCA13 CCA14 ## 0.0065 0.0062 0.0047 ## ## Eigenvalues for unconstrained axes:## CA1 CA2 CA3 CA4 CA5 CA6 CA7 CA8 CA9 ## 0.19776 0.14193 0.10117 0.07079 0.05330 0.03330 0.01887 0.01510 0.00949Redundancy Analysis
rda1 <- rda(varespec ~ ., data = varechem)rda1## Call: rda(formula = varespec ~ N + P + K + Ca + Mg + S + Al + Fe + Mn +## Zn + Mo + Baresoil + Humdepth + pH, data = varechem)## ## Inertia Proportion Rank## Total 1825.6594 1.0000 ## Constrained 1459.8891 0.7997 14## Unconstrained 365.7704 0.2003 9## Inertia is variance ## ## Eigenvalues for constrained axes:## RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7 RDA8 RDA9 RDA10 RDA11 RDA12 RDA13 ## 820.1 399.3 102.6 47.6 26.8 24.0 19.1 10.2 4.4 2.3 1.5 0.9 0.7 ## RDA14 ## 0.3 ## ## Eigenvalues for unconstrained axes:## PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 PC9 ## 186.19 88.46 38.19 18.40 12.84 10.55 5.52 4.52 1.09The cca.object
- Objects of class
"cca"are complex with many components - Entire class described in
?cca.object - Depending on what analysis performed some components may be
NULL - Used for (C)CA, PCA, RDA, CAP (
capscale()), and dbRDA (dbrda())
The cca.object
cca1 has a large number of components
$callhow the function was called$grand.totalin (C)CA sum ofrowsum$rowsumthe row sums$colsumthe column sums$tot.chitotal inertia, sum of Eigenvalues$pCCAConditioned (partial-ed out) components$CCAConstrained components$CAUnconstrained components$methodOrdination method used$inertiaDescription of what inertia is
The cca.object
Depending on how one called cca() etc some of these components will be NULL
$pCCA is only filled in if a partial constrained ordination fitted
rda() returns objects with classes "rda" and "cca", but in most cases those objects work like those of class "cca"
The Eigenvalues and axis scores are now spread about the $CA and $CCA components (also $pCCA if a partial CCA)
Thankfully we can use extractor functions to get at such things
Eigenvalues
Use eigenvals() to extract Eigenvalues from a fitted ordination object
eigenvals(cca1)## CCA1 CCA2 CCA3 CCA4 CCA5 CCA6 CCA7 CCA8 ## 0.4388704 0.2917753 0.1628465 0.1421302 0.1179519 0.0890291 0.0702945 0.0583592 ## CCA9 CCA10 CCA11 CCA12 CCA13 CCA14 CA1 CA2 ## 0.0311408 0.0132944 0.0083644 0.0065385 0.0061563 0.0047332 0.1977645 0.1419256 ## CA3 CA4 CA5 CA6 CA7 CA8 CA9 ## 0.1011741 0.0707868 0.0533034 0.0332994 0.0188676 0.0151044 0.0094876Example
- Fit a CCA model to the lichen pasture data. The model should include, N, P, and K only.
- Save the model in object
mycca1 - How much variance is explained by this model?
- Extract the eigenvalues, how many constrained axes are there?
library("vegan")data(varechem, varespec)library("vegan")data(varechem, varespec)mycca1 <- cca(varespec ~ N + P + K, data = varechem)mycca1## Call: cca(formula = varespec ~ N + P + K, data = varechem)## ## Inertia Proportion Rank## Total 2.0832 1.0000 ## Constrained 0.4464 0.2143 3## Unconstrained 1.6368 0.7857 20## Inertia is scaled Chi-square ## ## Eigenvalues for constrained axes:## CCA1 CCA2 CCA3 ## 0.19309 0.16271 0.09060 ## ## Eigenvalues for unconstrained axes:## CA1 CA2 CA3 CA4 CA5 CA6 CA7 CA8 ## 0.4495 0.2870 0.1877 0.1675 0.1280 0.1050 0.0750 0.0629 ## (Showing 8 of 20 unconstrained eigenvalues)ev <- eigenvals(mycca1, model = "constrained")head(ev)## CCA1 CCA2 CCA3 ## 0.19308594 0.16271109 0.09060228length(ev)## [1] 3Extracting axis scores
To extract a range of scores from a fitted ordination use scores()
- takes an ordination object as the first argument
choices— which axes? Defaults toc(1,2)display— which type(s) of scores to return"sites"or"wa": scores for samples in response matrix"species": scores for variables/columns in response"lc": linear combination site scores"bp": biplot scores (coords of arrow tip)"cn": centroid scores (coords of factor centroids)
Extracting axis scores
str(scores(cca1, choices = 1:4, display = c("species","sites")), max = 1)## List of 2## $ species: num [1:44, 1:4] 0.0753 -0.1813 -1.0535 -1.2774 -0.1526 ...## ..- attr(*, "dimnames")=List of 2## $ sites : num [1:24, 1:4] 0.178 -0.97 -1.28 -1.501 -0.598 ...## ..- attr(*, "dimnames")=List of 2head(scores(cca1, choices = 1:2, display = "sites"))## CCA1 CCA2## 18 0.1784733 -1.0598842## 15 -0.9702382 -0.1971387## 24 -1.2798478 0.4764498## 27 -1.5009195 0.6521559## 23 -0.5980933 -0.1840362## 19 -0.1102881 0.7143142Scalings…
When we draw the results of many ordinations we display 2 or more sets of data
Can't display all of these and maintain relationships between the scores
Solution scale one set of scores relative to the other via the scaling argument
Scalings…
scaling = 1— Focus on sites, scale site scores by λiλiscaling = 2— Focus on species, scale species scores by λiλiscaling = 3— Symmetric scaling, scale both scores by √λi√λiscaling = -1— As above, butscaling = -2— Forcca()multiply results by √(1/(1−λi))√(1/(1−λi))scaling = -3— this is Hill's scalingscaling < 0— Forrda()divide species scores by species' σσscaling = 0— raw scores
scores(cca1, choices = 1:2, display = "species", scaling = 3)Scalings…
Thankfully we can use alternative descrpitors to extract scores:
"none""sites""species""symmetric"
Two modifiers select negative scores depending on whether the model is CCA or RDA:
hill = TRUEcorrelation = TRUE
Example
- Using the CCA model you fitted, extract the site scores for axes 2 and 3 with Hill's scaling, focusing on the sites
Example
- Using the CCA model you fitted, extract the site scores for axes 2 and 3 with Hill's scaling, focusing on the sites
scrs <- scores(mycca1, display = "sites", choices = c(2,3), scaling = "sites", hill = TRUE)head(scrs)## CCA2 CCA3## 18 0.21507383 -0.22617222## 15 -0.53564592 -0.14736699## 24 -0.28328352 -0.56306912## 27 -0.79825273 -0.35205393## 23 -0.06029273 -0.09438971## 19 0.04742753 0.09586591Partial constrained ordinations
Partial constrained ordinations remove the effect of one or more variables then fit model of interest
Argument Z is used for a data frame of variables to partial out
pcca <- cca(X = varespec, Y = varechem[, "Ca", drop = FALSE], Z = varechem[, "pH", drop = FALSE])Or with the formula interface use the Condition() function
pcca <- cca(varespec ~ Ca + Condition(pH), data = varechem) ## easier!Partial constrained ordinations
pcca <- cca(varespec ~ Ca + Condition(pH), data = varechem) ## easier!pcca## Call: cca(formula = varespec ~ Ca + Condition(pH), data = varechem)## ## Inertia Proportion Rank## Total 2.0832 1.0000 ## Conditional 0.1458 0.0700 1## Constrained 0.1827 0.0877 1## Unconstrained 1.7547 0.8423 21## Inertia is scaled Chi-square ## ## Eigenvalues for constrained axes:## CCA1 ## 0.18269 ## ## Eigenvalues for unconstrained axes:## CA1 CA2 CA3 CA4 CA5 CA6 CA7 CA8 ## 0.3834 0.2749 0.2123 0.1760 0.1701 0.1161 0.1089 0.0880 ## (Showing 8 of 21 unconstrained eigenvalues)Triplots
Triplots will generally produce a mess; we can really only display a couple of bits approximately anyway Trying to cram three things in is a recipe for a mess… but we can do it
plot(cca1)Model building
Building constrained ordination models
If we don't want to think it's easy to fit a poor model with many constraints — That's what I did with cca1 and rda1
Remember, CCA and RDA are just regression methods — everything you know about regression applies here
A better approach is to think about the important variables and include only those
The formula interface allows you to create interaction or quadratic terms easily (though be careful with latter)
It also handles factor or class constraints automatically unlike the basic interface
Building constrained ordination models
vare.cca <- cca(varespec ~ Al + P*(K + Baresoil), data = varechem)vare.cca## Call: cca(formula = varespec ~ Al + P * (K + Baresoil), data =## varechem)## ## Inertia Proportion Rank## Total 2.083 1.000 ## Constrained 1.046 0.502 6## Unconstrained 1.038 0.498 17## Inertia is scaled Chi-square ## ## Eigenvalues for constrained axes:## CCA1 CCA2 CCA3 CCA4 CCA5 CCA6 ## 0.3756 0.2342 0.1407 0.1323 0.1068 0.0561 ## ## Eigenvalues for unconstrained axes:## CA1 CA2 CA3 CA4 CA5 CA6 CA7 CA8 ## 0.27577 0.15411 0.13536 0.11803 0.08887 0.05511 0.04919 0.03781 ## (Showing 8 of 17 unconstrained eigenvalues)Building constrained ordination models
For CCA, RDA etc we have little choice but to do
- Fit well-chosen set of candidate models & compare, or
- Fit a full model of well-chosen variables & then do stepwise selection
Automatic approaches to model building should be used cautiously!
The standard step() function can be used as vegan provides two helper methods, deviance() and extractAIC(), used by step()
vegan also provides methods for class "cca" for add1() and drop1()
Variance inflation factors
Linear dependencies between constraints can be investigated with VIF
VIF is a measure of how much the variance of ˆβj^βj is inflated by presence of other covariates
Lots of rules of thumb
- VIF >= 20 indicates strong collinearity in constraints
- VIF >= 10 potentially of concern & should be looked at
Computed via vif.cca()
vif.cca(cca1)## N P K Ca Mg S Al Fe ## 1.981742 6.028515 12.009357 9.925801 9.810609 18.378794 21.192739 9.127762 ## Mn Zn Mo Baresoil Humdepth pH ## 5.380432 7.739664 4.320346 2.253683 6.012537 7.389267Stepwise selection in CCA
step() uses AIC which is a fudge for RDA/CCA — use ordistep()
- Define an upper and lower model scope, say the full model and the null model
- To step from the lower scope or null model we use
upr <- cca(varespec ~ ., data = varechem)lwr <- cca(varespec ~ 1, data = varechem)set.seed(1)mods <- ordistep(lwr, scope = formula(upr), trace = 0)trace = 0 is used here to turn off printing of progress
Permutation tests are used (more on these later); the theory for an AIC for ordination is somewhat loose
Stepwise selection in CCA
The object returned by step() is a standard "cca" object with an extra component $anova
mods## Call: cca(formula = varespec ~ Al + P + K, data = varechem)## ## Inertia Proportion Rank## Total 2.0832 1.0000 ## Constrained 0.6441 0.3092 3## Unconstrained 1.4391 0.6908 20## Inertia is scaled Chi-square ## ## Eigenvalues for constrained axes:## CCA1 CCA2 CCA3 ## 0.3616 0.1700 0.1126 ## ## Eigenvalues for unconstrained axes:## CA1 CA2 CA3 CA4 CA5 CA6 CA7 CA8 ## 0.3500 0.2201 0.1851 0.1551 0.1351 0.1003 0.0773 0.0537 ## (Showing 8 of 20 unconstrained eigenvalues)Stepwise selection in CCA
The $anova component contains a summary of the steps involved in automatic model building
mods$anova## Df AIC F Pr(>F) ## + Al 1 128.61 3.6749 0.005 **## + P 1 127.91 2.5001 0.005 **## + K 1 127.44 2.1688 0.050 * ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Aladded first, thenP, followed byK, then stopped
Stepwise selection in CCA
Step-wise model selection is fairly fragile; if we start from the full model we won't end up with the same final model
mods2 <- step(upr, scope = list(lower = formula(lwr), upper = formula(upr)), trace = 0, test = "perm")mods2Stepwise selection in CCA
mods2 <- step(upr, scope = list(lower = formula(lwr), upper = formula(upr)), trace = 0, test = "perm")mods2## Call: cca(formula = varespec ~ P + K + Mg + S + Mn + Mo + Baresoil +## Humdepth, data = varechem)## ## Inertia Proportion Rank## Total 2.0832 1.0000 ## Constrained 1.1165 0.5360 8## Unconstrained 0.9667 0.4640 15## Inertia is scaled Chi-square ## ## Eigenvalues for constrained axes:## CCA1 CCA2 CCA3 CCA4 CCA5 CCA6 CCA7 CCA8 ## 0.4007 0.2488 0.1488 0.1266 0.0875 0.0661 0.0250 0.0130 ## ## Eigenvalues for unconstrained axes:## CA1 CA2 CA3 CA4 CA5 CA6 CA7 CA8 CA9 CA10 ## 0.25821 0.18813 0.11927 0.10204 0.08791 0.06085 0.04461 0.02782 0.02691 0.01646 ## CA11 CA12 CA13 CA14 CA15 ## 0.01364 0.00823 0.00655 0.00365 0.00238Adjusted R2R2 for ordination models
Ordinary R2R2 is biased for the same reasons as for a linear regression
- adding a variable to constraints will increase R2R2
- the larger the number of constraints in the model the larger R2R2, is due to random correlations
Can attempt to account for this bias via an adjusted R2R2 measure
Adjusted R2R2 for ordination models
Can attempt to account for this bias via an adjusted R2R2 measure
R2adj=1−n−1n−m−1(1−R2)R2adj=1−n−1n−m−1(1−R2)
- nn is number of samples mm is number of constraints (model degrees of freedom)
- Can be used up to ∼M>n/2∼M>n/2 before becomes too conservative
- Can be negative
RsquareAdj(cca1)## $r.squared## [1] 0.6919576## ## $adj.r.squared## [1] 0.2163163Stepwise selection via adjusted R2R2
Problems with stepwise selection are myriad. Affects RDA, CCA, etc
Blanchet et al (2008) proposed a two-step solution for models where R2adjR2adj makes sense
Stepwise selection via adjusted R2R2
- Global test of all constraints
- Proceed only if this test is significant
- Helps prevent inflation of overall type I error
- Proceed with forward selection, but with two stopping rules
- Usual significance threshold αα
- The global R2adjR2adj
- Stop if next candidate model is non-significant or if R2adjR2adj exceeds the global R2adjR2adj
Available in ordiR2step()
Stepwise selection via adjusted R2R2
ordiR2step(lwr, upr, trace = FALSE)## Call: cca(formula = varespec ~ Al + P + K, data = varechem)## ## Inertia Proportion Rank## Total 2.0832 1.0000 ## Constrained 0.6441 0.3092 3## Unconstrained 1.4391 0.6908 20## Inertia is scaled Chi-square ## ## Eigenvalues for constrained axes:## CCA1 CCA2 CCA3 ## 0.3616 0.1700 0.1126 ## ## Eigenvalues for unconstrained axes:## CA1 CA2 CA3 CA4 CA5 CA6 CA7 CA8 ## 0.3500 0.2201 0.1851 0.1551 0.1351 0.1003 0.0773 0.0537 ## (Showing 8 of 20 unconstrained eigenvalues)Permutation tests
Permutation tests in vegan
RDA has lots of theory behind it, CCA bit less. However, ecological/environmental data invariably violate what little theory we have
Instead we use permutation tests to assess the importance of fitted models — the data are shuffled in some way and the model refitted to derive a Null distribution under some hypothesis of no effect
Permutation tests in vegan
What is shuffled and how is of paramount importance for the test to be valid
- No conditioning (partial) variables then rows of the species data are permuted
- With conditioning variables, two options are available, both of which permute residuals from model fits
- The full model uses residuals from model Y=X+Z+εY=X+Z+ε
- The reduced model uses residuals from model Y=Z+εY=Z+ε
- In vegan which is used can be set via argument
modelwith"direct","full", and"reduced"respectively
Permutation tests in vegan
A test statistic is required, computed for observed model & each permuted model
vegan uses a pseudo FF statistic
F=χ2model/dfmodelχ2resid/dfresidF=χ2model/dfmodelχ2resid/dfresid
Evaluate whether FF is unusually large relative to the null (permutation) distribution of FF
Permutation tests in vegan
pstat <- permustats(anova(cca1))summary(pstat)## ## statistic SES mean lower median upper Pr(perm) ## Model 1.4441 2.1270 1.0215 1.0033 1.3824 0.035 *## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## ## (Interval (Upper - Lower) = 0.95)densityplot(pstat)The summary method of permustats estimates the standardized effect sizes (SES) as the difference of observed statistic and mean of permutations divided by the standard deviation of permutations (also known as z-values). It also prints the the mean, median, and limits which contain interval percent of permuted values. With the default (interval = 0.95), for two-sided test these are (2.5%, 97.5%) and for one-sided tests either 5% or 95% quantile and the p-value depending on the test direction. The mean, quantiles and z values are evaluated from permuted values without observed statistic, but the p-value is evaluated with the observed statistic.
Permutation tests in vegan: anova()
- The main user function is the
anova()method - It is an interface to the lower-level function
permutest.cca() - At its most simplest, the
anova()method tests whether the model as a whole is significant
Permutation tests in vegan: anova()
F=1.4415/140.6417/9=1.4441F=1.4415/140.6417/9=1.4441
set.seed(42)(perm <- anova(cca1))## Permutation test for cca under reduced model## Permutation: free## Number of permutations: 999## ## Model: cca(formula = varespec ~ N + P + K + Ca + Mg + S + Al + Fe + Mn + Zn + Mo + Baresoil + Humdepth + pH, data = varechem)## Df ChiSquare F Pr(>F) ## Model 14 1.44148 1.4441 0.029 *## Residual 9 0.64171 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Permutation tests in vegan: anova()
anova.cca() has a number of arguments
args(anova.cca)## function (object, ..., permutations = how(nperm = 999), by = NULL, ## model = c("reduced", "direct", "full"), parallel = getOption("mc.cores"), ## strata = NULL, cutoff = 1, scope = NULL) ## NULLobject is the fitted ordination
permutations controls what is permuted and how
by determines what is tested; the default is to test the model
Types of permutation test in vegan
A number of types of test can be envisaged
- Testing the overall significance of the model
- Testing constrained (canonical) axes
- Testing individual model terms sequentially
- The marginal effect of a single variable
The first is the default in anova()
The other three can be selected via the argument by
Testing canonical axes
- The constrained (canonical) axes can be individually tests by specifying
by = "axis" - The first axis is tested in terms of variance explained compared to residual variance
- The second axis is tested after partialling out the first axis…
- and so on
Testing canonical axes
set.seed(1)anova(mods, by = "axis")## Permutation test for cca under reduced model## Forward tests for axes## Permutation: free## Number of permutations: 999## ## Model: cca(formula = varespec ~ Al + P + K, data = varechem)## Df ChiSquare F Pr(>F) ## CCA1 1 0.36156 5.0249 0.001 ***## CCA2 1 0.16996 2.3621 0.034 * ## CCA3 1 0.11262 1.5651 0.142 ## Residual 20 1.43906 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Testing terms sequentially
- The individual terms in the model can be tested using
by = "terms" - The terms are assessed in the order they were specified in the model, sequentially from first to last
- Test is of the additional variance explained by adding the kkth variable to the model
- Ordering of the terms will affect the results
Testing terms sequentially
set.seed(5)anova(mods, by = "terms")## Permutation test for cca under reduced model## Terms added sequentially (first to last)## Permutation: free## Number of permutations: 999## ## Model: cca(formula = varespec ~ Al + P + K, data = varechem)## Df ChiSquare F Pr(>F) ## Al 1 0.29817 4.1440 0.001 ***## P 1 0.18991 2.6393 0.005 ** ## K 1 0.15605 2.1688 0.018 * ## Residual 20 1.43906 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Testing terms marginal effects
- The marginal effect of a model term can be assessed using
by = "margin" - The marginal effect is the effect of a particular term when all other model terms are included in the model
Testing terms marginal effects
set.seed(10)anova(mods, by = "margin")## Permutation test for cca under reduced model## Marginal effects of terms## Permutation: free## Number of permutations: 999## ## Model: cca(formula = varespec ~ Al + P + K, data = varechem)## Df ChiSquare F Pr(>F) ## Al 1 0.31184 4.3340 0.001 ***## P 1 0.16810 2.3362 0.016 * ## K 1 0.15605 2.1688 0.029 * ## Residual 20 1.43906 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Spring meadow vegetation
Example & data taken from Leps & Smilauer (2014), Case Study 2
Spring fen meadow vegetation in westernmost Carpathian mountains
## load veganlibrary("vegan")## load the dataspp <- read.csv("data/meadow-spp.csv", header = TRUE, row.names = 1)env <- read.csv("data/meadow-env.csv", header = TRUE, row.names = 1)Spring meadow vegetation
CCA a reasonable starting point as the gradient is long here (check with decorana() if you want)
m1 <- cca(spp ~ ., data = env)set.seed(32)anova(m1)## Permutation test for cca under reduced model## Permutation: free## Number of permutations: 999## ## Model: cca(formula = spp ~ Ca + Mg + Fe + K + Na + Si + SO4 + PO4 + NO3 + NH3 + Cl + Corg + pH + conduct + slope, data = env)## Df ChiSquare F Pr(>F) ## Model 15 1.5597 1.497 0.001 ***## Residual 54 3.7509 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Spring meadow vegetation
plot(m1)Spring meadow vegetation
set.seed(67)lwr <- cca(spp ~ 1, data = env)( m2 <- ordistep(lwr, scope = formula(m1), trace = FALSE) )## Call: cca(formula = spp ~ Ca + conduct + Corg + Na + NH3 + Fe + pH,## data = env)## ## Inertia Proportion Rank## Total 5.3107 1.0000 ## Constrained 0.9899 0.1864 7## Unconstrained 4.3208 0.8136 62## Inertia is scaled Chi-square ## ## Eigenvalues for constrained axes:## CCA1 CCA2 CCA3 CCA4 CCA5 CCA6 CCA7 ## 0.4268 0.1447 0.1116 0.0936 0.0760 0.0719 0.0652 ## ## Eigenvalues for unconstrained axes:## CA1 CA2 CA3 CA4 CA5 CA6 CA7 CA8 ## 0.27251 0.19518 0.16703 0.14993 0.14606 0.14168 0.13292 0.12154 ## (Showing 8 of 62 unconstrained eigenvalues)Spring meadow vegetation
plot(m2)Spring meadow vegetation
m2$anova## Df AIC F Pr(>F) ## + Ca 1 453.14 4.7893 0.005 **## + conduct 1 453.29 1.7915 0.005 **## + Corg 1 453.61 1.6011 0.005 **## + Na 1 453.93 1.5827 0.010 **## + NH3 1 454.36 1.4507 0.020 * ## + Fe 1 454.89 1.3386 0.040 * ## + pH 1 455.46 1.2756 0.040 * ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Spring meadow vegetation
Alternative is RDA with a transformation
spph <- decostand(spp, method = "hellinger")m3 <- rda(spph ~ ., data = env)lwr <- rda(spph ~ 1, data = env)m4 <- ordistep(lwr, scope = formula(m3), trace = FALSE)m4## Call: rda(formula = spph ~ Ca + NH3 + conduct + Si + Corg + NO3 + pH +## Mg, data = env)## ## Inertia Proportion Rank## Total 0.6123 1.0000 ## Constrained 0.1823 0.2977 8## Unconstrained 0.4300 0.7023 61## Inertia is variance ## ## Eigenvalues for constrained axes:## RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7 RDA8 ## 0.10572 0.02148 0.01224 0.01148 0.00945 0.00891 0.00696 0.00609 ## ## Eigenvalues for unconstrained axes:## PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 ## 0.04311 0.03026 0.02030 0.01767 0.01649 0.01519 0.01383 0.01346 ## (Showing 8 of 61 unconstrained eigenvalues)Spring meadow vegetation
plot(m4)Spring meadow vegetation
Stepwise using R2adj
m5 <- ordiR2step(lwr, scope = formula(m3), trace = FALSE)m5$anova## R2.adj Df AIC F Pr(>F) ## + Ca 0.12588 1 -41.779 10.9370 0.002 **## + NH3 0.14628 1 -42.468 2.6242 0.002 **## + conduct 0.16322 1 -42.925 2.3570 0.002 **## + Si 0.17711 1 -43.164 2.1136 0.002 **## + Corg 0.18518 1 -42.940 1.6442 0.014 * ## + NO3 0.19257 1 -42.680 1.5853 0.010 **## + pH 0.19966 1 -42.417 1.5583 0.012 * ## <All variables> 0.20332 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Restricted permutation tests
Restricted permutation tests
What is shuffled and how is of paramount importance for a valid test
Complete randomisation assumes a null hypothesis where all observations are independent
- Temporal or spatial correlation
- Clustering, repeated measures
- Nested sampling designs (Split-plots designs)
- Blocks
- …
Permutation must give null distribution of the test statistic whilst preserving the dependence between observations
Trick is to shuffle the data whilst preserving that dependence
Restricted permutations
Canoco has had restricted permutations for a long time. vegan has only recently caught up & we're not (quite) there yet
vegan used to only know how to completely randomise data or completely randomise within blocks (via strata in vegan)
The permute package grew out of initial code in the vegan repository to generate the sorts of restricted permutations available in Canoco
We have now fully integrated permute into vegan…
vegan depends on permute so it will already be installed & loaded when using vegan
Restricted permutations with permute
permute follows Canoco closely — at the (friendly!) chiding of Cajo ter Braak when it didn't do what he wanted!
Samples can be thought of as belonging to three levels of a hierarchy
- the sample level; how are individual samples permuted
- the plot level; how are samples grouped at an intermediate level
- the block level; how are samples grouped at the outermost level
Blocks define groups of plots, each of which can contain groups of samples
Restricted permutations with permute
Blocks are never permuted; if defined, only plots or samples within the blocks get shuffled & samples are never swapped between blocks
Plots or samples within plots, or both can be permuted following one of four simple permutation types
- Free permutation (randomisation)
- Time series or linear transect, equal spacing
- Spatial grid designs, equal regular spacing
- Permutation of plots (groups of samples)
- Fixed (no permutation)
Multiple plots per block, multiple samples per plot; plots could be arranged in a spatial grid & samples within plots form time series
Blocks
Blocks are a random factor that does not interact with factors that vary within blocks
Blocks form groups of samples that are never permuted between blocks, only within blocks
Using blocks you can achieve what the strata argument used to in vegan; needs to be a factor variable
The variation between blocks should be excluded from the test; permute doesn't do this for you!
Use + Condition(blocks) in the model formula where blocks is a factor containing the block membership for each observation
Time series & linear transects
Can link randomly starting point of one series to any time point of another series if series are stationary under H0 that series are unrelated
Achieve this via cyclic shift permutations — wrap series into a circle
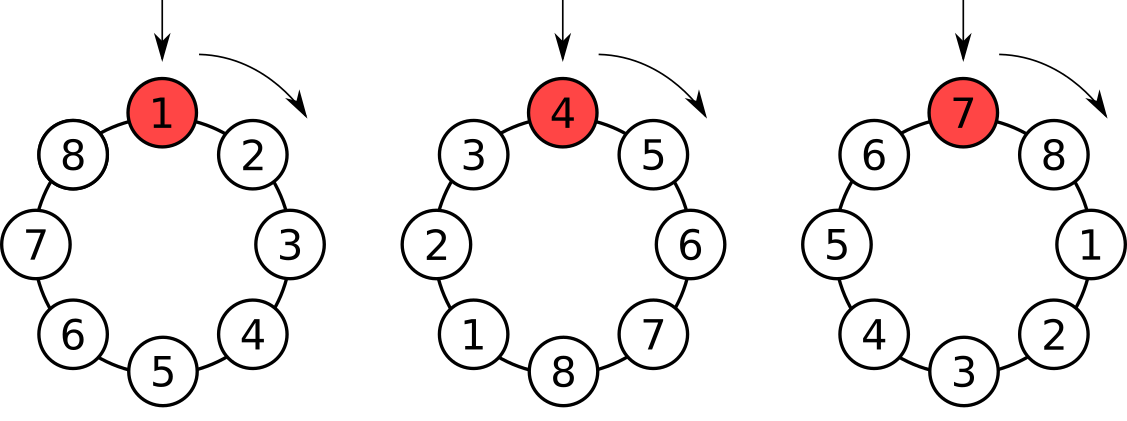
Time series & linear transects
Works OK if there are no trends or cyclic pattern — autocorrelation structure only broken at the end points if series are stationary
Can detrend to make series stationary but not if you want to test significance of a trend
shuffle(10, control = how(within = Within(type = "series")))## [1] 6 7 8 9 10 1 2 3 4 5Spatial grids
The trick of cyclic shifts can be extended to two dimensions for a regular spatial grid arrangement of points
Now shifts are toroidal as we join the end point in the x direction together and in the y direction together
Source: Dave Burke, Wikimedia CC BY
set.seed(4)h <- how(within = Within(type = "grid", ncol = 3, nrow = 3))perm <- shuffle(9, control = h)matrix(perm, ncol = 3)## [,1] [,2] [,3]## [1,] 1 4 7## [2,] 2 5 8## [3,] 3 6 9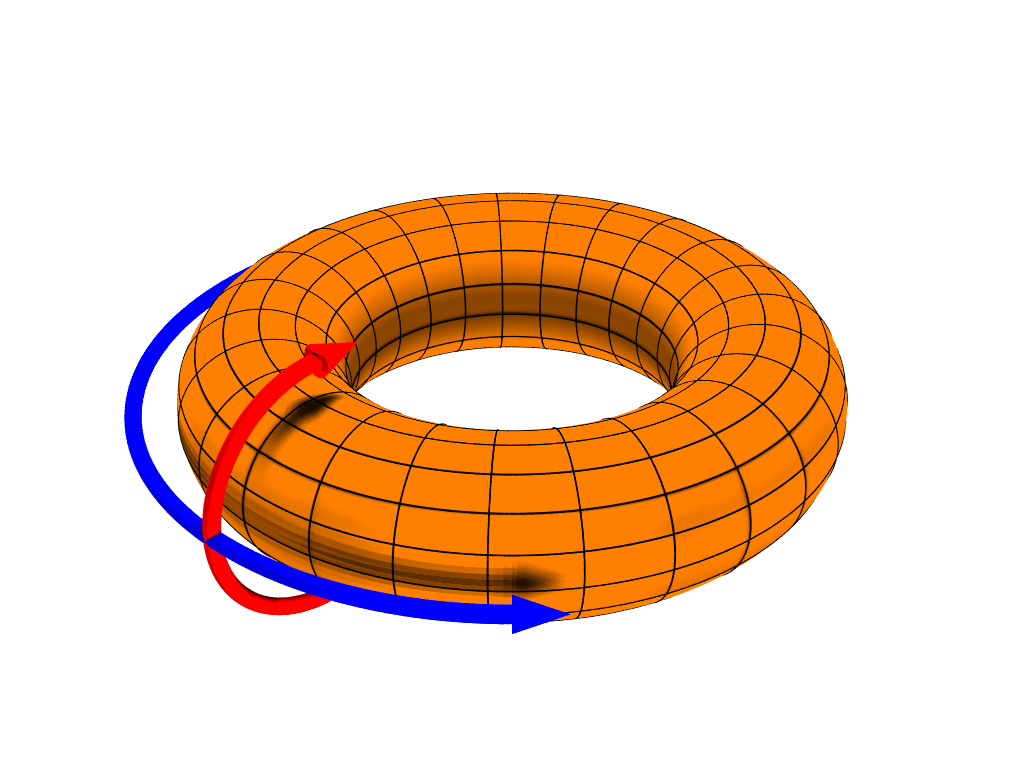
Whole-plots & split-plots I
Split-plot designs are hierarchical with two levels of units
- whole-plots , which contain
- split-plots (the samples)
Permute one or both, but whole-plots must be of equal size
Essentially allows more than one error stratum to be analyzed
Test effect of constraints that vary between whole plots by permuting the whole-plots whilst retaining order of split-splots (samples) within the whole-plots
Test effect of constraints that vary within whole-plots by permuting the split-plots within whole-plots without permuting the whole-plots
Whole-plots & split-plots II
Whole-plots or split-plots, or both, can be time series, linear transects or rectangular grids in which case the appropriate restricted permutation is used
If the split-plots are parallel time series & time is an autocorrelated error component affecting all series then the same cyclic shift can be applied to each time series (within each whole-plot) (constant = TRUE)
Mirrored permutations
Mirroring in restricted permutations allows for isotropy in dependencies by reflecting the ordering of samples in time or spatial dimensions
For a linear transect, technically the autocorrelation at lag h is equal to that at lag -h (also in a trend-free time series)
Mirrored permutations
Hence the series (1, 2, 3, 4) and (4, 3, 2, 1) are equivalent fom this point of view & we can draw permutations from either version
Similar argument can be made for spatial grids
Using mirror = TRUE then can double (time series, linear transects) or quadruple (spatial grids) the size of the set of permutations
Sets of permutations — no free lunch
Restricted severely reduce the size of the set of permutations
As the minimum p value obtainable is 1/np where np is number of allowed permutations (including the observed) this can impact the ability to detect signal/pattern
If we don't want mirroring
- in a time series of 20 samples the minimum p is 1/20 (0.05)
- in a time series of 100 samples the minimum p is 1/100 (0.01)
- in a data set with 10 time series each of 20 observations (200 total), if we assume an autocorrelated error component over all series (
constant = TRUE) then there are only 20 permutations of the data and minimum p is 0.05
Sets of permutations — no free lunch
When the set of permutations is small it is better to switch to an exact test & evaluate all permutations in the set rather than randomly sample from the set
Use complete = TRUE in the call to how() — perhaps also increase maxperm
Designing permutation schemes
In permute, we set up a permutation scheme with how()
We sample from the permutation scheme with
shuffle(), which gives a single draw from scheme, orshuffleSet(), which returns a set ofndraws from the scheme
allPerms() can generated the entire set of permutations — note this was designed for small sets of permutations & is slow if you request it for a scheme with many thousands of permutations!
Designing permutation schemes
how() has three main arguments
within— takes input from helperWithin()plots— takes input from helperPlots()blocks— takes a factor variable as input
plt <- gl(3, 10)h <- how(within = Within(type = "series"), plots = Plots(strata = plt))Designing permutation schemes
Helper functions make it easy to change one or a few aspects of permutation scheme, rest left at defaults
args(Within)## function (type = c("free", "series", "grid", "none"), constant = FALSE, ## mirror = FALSE, ncol = NULL, nrow = NULL) ## NULLargs(Plots)## function (strata = NULL, type = c("none", "free", "series", "grid"), ## mirror = FALSE, ncol = NULL, nrow = NULL) ## NULLDesigning permutation schemes
how() has additional arguments, many of which control the heuristics that kick in to stop you shooting yourself in the foot and demanding 9999 permutations when there are only 10
completeshould we enumerate the entire set of permutations?minpermlower bound on the size of the set of permutations at & below which we turn on complete enumeration
args(how)## function (within = Within(), plots = Plots(), blocks = NULL, ## nperm = 199, complete = FALSE, maxperm = 9999, minperm = 5040, ## all.perms = NULL, make = TRUE, observed = FALSE) ## NULLTime series example I
Time series within 3 plots, 10 observation each
plt <- gl(3, 10)h <- how(within = Within(type = "series"), plots = Plots(strata = plt))set.seed(4)p <- shuffle(30, control = h)do.call("rbind", split(p, plt)) ## look at perms in context## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]## 1 9 10 1 2 3 4 5 6 7 8## 2 14 15 16 17 18 19 20 11 12 13## 3 24 25 26 27 28 29 30 21 22 23Time series example II
Time series within 3 plots, 10 observation each, same permutation within each
plt <- gl(3, 10)h <- how(within = Within(type = "series", constant = TRUE), plots = Plots(strata = plt))set.seed(4)p <- shuffle(30, control = h)do.call("rbind", split(p, plt)) ## look at perms in context## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]## 1 9 10 1 2 3 4 5 6 7 8## 2 19 20 11 12 13 14 15 16 17 18## 3 29 30 21 22 23 24 25 26 27 28Ohraz Case Study
Restricted permutations | Ohraz
Now we've seen how to drive permute, we can use the same how() commands to set up permutation designs within vegan functions
Analyse the Ohraz data Case study 5 of Leps & Smilauer
Repeated observations of composition from an experiment
- Factorial design (3 replicates)
- Treatments: fertilisation, mowing, Molinia removal
Test 1 of the hypotheses
There are no directional changes in species composition in time that are common to all treatments or specific treatments
Restricted permutations | Ohraz
Analyse the Ohraz data Case study 5 of Leps & Smilauer
## load veganlibrary("vegan")## load the dataspp <- read.csv("data/ohraz-spp.csv", header = TRUE, row.names = 1)env <- read.csv("data/ohraz-env.csv", header = TRUE, row.names = 1)molinia <- spp[, 1]spp <- spp[, -1]## Year as numericenv <- transform(env, year = as.numeric(as.character(year)))Restricted permutations | Ohraz
c1 <- rda(spp ~ year + year:mowing + year:fertilizer + year:removal + Condition(plotid), data = env)(h <- how(within = Within(type = "none"), plots = Plots(strata = env$plotid, type = "free")))## ## Permutation Design:## ## Blocks:## Defined by: none## ## Plots:## Plots: env$plotid## Permutation type: free## Mirrored?: No## ## Within Plots:## Permutation type: none## ## Permutation details:## Number of permutations: 199## Max. number of permutations allowed: 9999## Evaluate all permutations?: No. Activation limit: 5040Restricted permutations | Ohraz
set.seed(42)anova(c1, permutations = h, model = "reduced")## Permutation test for rda under reduced model## Plots: env$plotid, plot permutation: free## Permutation: none## Number of permutations: 199## ## Model: rda(formula = spp ~ year + year:mowing + year:fertilizer + year:removal + Condition(plotid), data = env)## Df Variance F Pr(>F) ## Model 4 158.85 6.4247 0.005 **## Residual 90 556.30 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Restricted permutations | Ohraz
set.seed(24)anova(c1, permutations = h, model = "reduced", by = "axis")## Permutation test for rda under reduced model## Forward tests for axes## Plots: env$plotid, plot permutation: free## Permutation: none## Number of permutations: 199## ## Model: rda(formula = spp ~ year + year:mowing + year:fertilizer + year:removal + Condition(plotid), data = env)## Df Variance F Pr(>F) ## RDA1 1 89.12 14.4173 0.005 **## RDA2 1 34.28 5.5458 0.005 **## RDA3 1 26.52 4.2900 0.025 * ## RDA4 1 8.94 1.4458 0.485 ## Residual 90 556.30 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Hierarchical analysis of crayfish
Hierarchical analysis of crayfish
Variation in communities may exist at various scales, sometimes hierarchically
A first step in understanding this variation is to test for its exisistence
In this example from Leps & Smilauer (2014) uses crayfish data from Spring River, Arkansas/Missouri, USA, collected by Dr. Camille Flinders.
567 records of 5 species, each sub-divided into Large & Small individuals
Hierarchical analysis of crayfish
## load datacrayfish <- head(read.csv("data/crayfish-spp.csv")[, -1], -1)design <- read.csv("data/crayfish-design.csv", skip = 1)[, -1]## fixup the namesnames(crayfish) <- gsub("\\.", "", names(crayfish))names(design) <- c("Watershed", "Stream", "Reach", "Run", "Stream.Nested", "ReachNested", "Run.Nested")Crayfish — Unconstrained
A number of samples have 0 crayfish, which excludes unimodal methods
m.pca <- rda(crayfish)summary(eigenvals(m.pca))## Importance of components:## PC1 PC2 PC3 PC4 PC5 PC6 PC7## Eigenvalue 3.5728 1.8007 1.1974 0.9012 0.79337 0.38886 0.28132## Proportion Explained 0.3818 0.1924 0.1280 0.0963 0.08478 0.04155 0.03006## Cumulative Proportion 0.3818 0.5742 0.7022 0.7985 0.88325 0.92480 0.95486## PC8 PC9 PC10## Eigenvalue 0.21225 0.20528 0.0048809## Proportion Explained 0.02268 0.02194 0.0005216## Cumulative Proportion 0.97754 0.99948 1.0000000Crayfish — Unconstrained
layout(matrix(1:2, ncol = 2))biplot(m.pca, type = c("text", "points"), scaling = "species")set.seed(23)ev.pca <- envfit(m.pca ~ Watershed, data = design, scaling = "species")plot(ev.pca, labels = levels(design$Watershed), add = FALSE)layout(1)Crayfish — Watershed scale
m.ws <- rda(crayfish ~ Watershed, data = design)m.ws## Call: rda(formula = crayfish ~ Watershed, data = design)## ## Inertia Proportion Rank## Total 9.3580 1.0000 ## Constrained 1.7669 0.1888 6## Unconstrained 7.5911 0.8112 10## Inertia is variance ## ## Eigenvalues for constrained axes:## RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 ## 0.7011 0.5540 0.3660 0.1064 0.0381 0.0013 ## ## Eigenvalues for unconstrained axes:## PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 PC9 PC10 ## 3.0957 1.2109 0.9717 0.7219 0.5333 0.3838 0.2772 0.2040 0.1879 0.0048Crayfish — Watershed scale
summary(eigenvals(m.ws, constrained = TRUE))## Importance of components:## RDA1 RDA2 RDA3 RDA4 RDA5 RDA6## Eigenvalue 0.7011 0.5540 0.3660 0.1064 0.03814 0.0012791## Proportion Explained 0.3968 0.3135 0.2072 0.0602 0.02159 0.0007239## Cumulative Proportion 0.3968 0.7103 0.9175 0.9777 0.99928 1.0000000Crayfish — Watershed scale
set.seed(1)ctrl <- how(nperm = 499, within = Within(type = "none"), plots = with(design, Plots(strata = Stream, type = "free")))(sig.ws <- anova(m.ws, permutations = ctrl))## Permutation test for rda under reduced model## Plots: Stream, plot permutation: free## Permutation: none## Number of permutations: 499## ## Model: rda(formula = crayfish ~ Watershed, data = design)## Df Variance F Pr(>F) ## Model 6 1.7669 21.724 0.002 **## Residual 560 7.5911 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Crayfish — Stream scale
m.str <- rda(crayfish ~ Stream + Condition(Watershed), data = design)m.str## Call: rda(formula = crayfish ~ Stream + Condition(Watershed), data =## design)## ## Inertia Proportion Rank## Total 9.3580 1.0000 ## Conditional 1.7669 0.1888 6## Constrained 1.1478 0.1227 10## Unconstrained 6.4433 0.6885 10## Inertia is variance ## Some constraints were aliased because they were collinear (redundant)## ## Eigenvalues for constrained axes:## RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7 RDA8 RDA9 RDA10 ## 0.4928 0.2990 0.2058 0.0782 0.0372 0.0224 0.0063 0.0030 0.0029 0.0002 ## ## Eigenvalues for unconstrained axes:## PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 PC9 PC10 ## 2.7853 0.8528 0.7737 0.6317 0.5144 0.2808 0.2517 0.1923 0.1559 0.0046Crayfish — Stream scale
summary(eigenvals(m.str, constrained = TRUE))## Importance of components:## RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7## Eigenvalue 0.4928 0.2990 0.2058 0.07824 0.03719 0.02235 0.006326## Proportion Explained 0.4293 0.2605 0.1793 0.06816 0.03240 0.01947 0.005511## Cumulative Proportion 0.4293 0.6898 0.8691 0.93731 0.96971 0.98918 0.994694## RDA8 RDA9 RDA10## Eigenvalue 0.003042 0.002894 0.0001546## Proportion Explained 0.002651 0.002521 0.0001347## Cumulative Proportion 0.997344 0.999865 1.0000000Crayfish — Stream scale
set.seed(1)ctrl <- how(nperm = 499, within = Within(type = "none"), plots = with(design, Plots(strata = Reach, type = "free")), blocks = with(design, Watershed))(sig.str <- anova(m.str, permutations = ctrl))## Permutation test for rda under reduced model## Blocks: with(design, Watershed) ## Plots: Reach, plot permutation: free## Permutation: none## Number of permutations: 499## ## Model: rda(formula = crayfish ~ Stream + Condition(Watershed), data = design)## Df Variance F Pr(>F) ## Model 14 1.1478 6.9477 0.004 **## Residual 546 6.4433 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Crayfish — Reach scale
(m.re <- rda(crayfish ~ Reach + Condition(Stream), data = design))## Call: rda(formula = crayfish ~ Reach + Condition(Stream), data =## design)## ## Inertia Proportion Rank## Total 9.3580 1.0000 ## Conditional 2.9148 0.3115 20## Constrained 1.4829 0.1585 10## Unconstrained 4.9603 0.5301 10## Inertia is variance ## Some constraints were aliased because they were collinear (redundant)## ## Eigenvalues for constrained axes:## RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7 RDA8 RDA9 RDA10 ## 0.6292 0.2706 0.2146 0.1414 0.1123 0.0467 0.0344 0.0270 0.0064 0.0003 ## ## Eigenvalues for unconstrained axes:## PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 PC9 PC10 ## 2.1635 0.6080 0.5605 0.5166 0.3749 0.2212 0.2052 0.1588 0.1477 0.0040Crayfish — Reach scale
set.seed(1)ctrl <- how(nperm = 499, within = Within(type = "none"), plots = with(design, Plots(strata = Run, type = "free")), blocks = with(design, Stream))(sig.re <- anova(m.re, permutations = ctrl))## Permutation test for rda under reduced model## Blocks: with(design, Stream) ## Plots: Run, plot permutation: free## Permutation: none## Number of permutations: 499## ## Model: rda(formula = crayfish ~ Reach + Condition(Stream), data = design)## Df Variance F Pr(>F) ## Model 42 1.4829 3.5875 0.002 **## Residual 504 4.9603 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Crayfish — Run scale
(m.run <- rda(crayfish ~ Run + Condition(Reach), data = design))## Call: rda(formula = crayfish ~ Run + Condition(Reach), data = design)## ## Inertia Proportion Rank## Total 9.3580 1.0000 ## Conditional 4.3977 0.4699 62## Constrained 1.8225 0.1948 10## Unconstrained 3.1378 0.3353 10## Inertia is variance ## Some constraints were aliased because they were collinear (redundant)## ## Eigenvalues for constrained axes:## RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7 RDA8 RDA9 RDA10 ## 0.8541 0.3141 0.1679 0.1393 0.1328 0.0835 0.0474 0.0429 0.0390 0.0016 ## ## Eigenvalues for unconstrained axes:## PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 PC9 PC10 ## 1.3137 0.4165 0.3832 0.2759 0.2378 0.1725 0.1215 0.1130 0.1016 0.0021Crayfish — Run scale
set.seed(1)ctrl <- how(nperm = 499, within = Within(type = "free"), blocks = with(design, Reach))(sig.run <- anova(m.run, permutations = ctrl))## Permutation test for rda under reduced model## Blocks: with(design, Reach) ## Permutation: free## Number of permutations: 499## ## Model: rda(formula = crayfish ~ Run + Condition(Reach), data = design)## Df Variance F Pr(>F) ## Model 126 1.8225 1.7425 0.002 **## Residual 378 3.1378 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1PERMANOVA
MANOVA
MANOVA is the multivariate form of ANOVA
- Multivariate response data
- Categorical predictor variables
Decompose variation in the responses into
- variation within groups
- variation between groups
Test to see if two is unusually large relative to H0
PERMANOVA
Doing that test requires lots of assumptions that rarely hold for ecological data
PERMANOVA: Permutational multivariate analysis of variance
Avoids most of these issues through the use of permutation tests
Directly decomposes a dissimilarity matrix into
- variation within groups
- variation between groups
PERMANOVA sensu stricto
vegan has four different ways to do essentially do this kind of analysis
adonis()— implements Anderson (2001)adonis2()— implements McArdle & Anderson (2001)dbrda()— implementation based on McArdle & Anderson (2001)capscale()— implements Legendre & Anderson (1999)
Be careful with adonis() as it allows only sequential tests
A difference between the functions is how they treat negative eigenvalues
The PERMANOVA idea
PERMANOA — adonis2()
data(dune, dune.env)adonis2(dune ~ Management*A1, data = dune.env, by = "terms")## Permutation test for adonis under reduced model## Terms added sequentially (first to last)## Permutation: free## Number of permutations: 999## ## adonis2(formula = dune ~ Management * A1, data = dune.env, by = "terms")## Df SumOfSqs R2 F Pr(>F) ## Management 3 1.4686 0.34161 3.2629 0.002 **## A1 1 0.4409 0.10256 2.9387 0.020 * ## Management:A1 3 0.5892 0.13705 1.3090 0.210 ## Residual 12 1.8004 0.41878 ## Total 19 4.2990 1.00000 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1PERMANOA — adonis2()
data(dune, dune.env)adonis2(dune ~ A1*Management, data = dune.env, by = "terms")## Permutation test for adonis under reduced model## Terms added sequentially (first to last)## Permutation: free## Number of permutations: 999## ## adonis2(formula = dune ~ A1 * Management, data = dune.env, by = "terms")## Df SumOfSqs R2 F Pr(>F) ## A1 1 0.7230 0.16817 4.8187 0.001 ***## Management 3 1.1865 0.27600 2.6362 0.005 ** ## A1:Management 3 0.5892 0.13705 1.3090 0.201 ## Residual 12 1.8004 0.41878 ## Total 19 4.2990 1.00000 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1PERMANOA — adonis2()
data(dune, dune.env)adonis2(dune ~ Management*A1, data = dune.env, by = "margin")## Permutation test for adonis under reduced model## Marginal effects of terms## Permutation: free## Number of permutations: 999## ## adonis2(formula = dune ~ Management * A1, data = dune.env, by = "margin")## Df SumOfSqs R2 F Pr(>F)## Management:A1 3 0.5892 0.13705 1.309 0.201## Residual 12 1.8004 0.41878 ## Total 19 4.2990 1.00000PERMANOA — adonis2()
data(dune, dune.env)adonis2(dune ~ Management*A1, data = dune.env, by = "margin")## Permutation test for adonis under reduced model## Marginal effects of terms## Permutation: free## Number of permutations: 999## ## adonis2(formula = dune ~ Management * A1, data = dune.env, by = "margin")## Df SumOfSqs R2 F Pr(>F)## Management:A1 3 0.5892 0.13705 1.309 0.201## Residual 12 1.8004 0.41878 ## Total 19 4.2990 1.00000The interaction is the only term that isn't marginal to other terms; not significant
PERMANOA — adonis2()
adonis2(dune ~ Management + A1, data = dune.env, by = "margin")## Permutation test for adonis under reduced model## Marginal effects of terms## Permutation: free## Number of permutations: 999## ## adonis2(formula = dune ~ Management + A1, data = dune.env, by = "margin")## Df SumOfSqs R2 F Pr(>F) ## Management 3 1.1865 0.27600 2.4828 0.008 **## A1 1 0.4409 0.10256 2.7676 0.020 * ## Residual 15 2.3895 0.55583 ## Total 19 4.2990 1.00000 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The dispersion problem
Anderson (2001) noted that PERMANOVA could confound location & dispersion effects
If one or more groups are more variable — dispersed around the centroid — than the others, this can result in a false detection of a difference of means — a location effect
Same problem affects t tests, ANOVA
Warton et al (2012) Anderson & Walsh (2013) Anderson et al (2017)
Dispersion
Test for dispersion effects
Marti Anderson (2006) developed a test for multivariate dispersions — PERMDISP2
- Calculate how far each observation is from its group median (or centroid)
- Take the absolute values of these distances-to-medians
- Do an ANOVA on the absolute distances with the groups as covariates
- Test the H0 of equal absolute distances to median among groups using a permutation test
In vegan this is betadisper()
Test for dispersion effects
data(varespec)dis <- vegdist(varespec) # Bray-Curtis distances## First 16 sites grazed, remaining 8 sites ungrazedgroups <- factor(c(rep(1,16), rep(2,8)), labels = c("grazed","ungrazed"))mod <- betadisper(dis, groups)mod## ## Homogeneity of multivariate dispersions## ## Call: betadisper(d = dis, group = groups)## ## No. of Positive Eigenvalues: 15## No. of Negative Eigenvalues: 8## ## Average distance to median:## grazed ungrazed ## 0.3926 0.2706 ## ## Eigenvalues for PCoA axes:## (Showing 8 of 23 eigenvalues)## PCoA1 PCoA2 PCoA3 PCoA4 PCoA5 PCoA6 PCoA7 PCoA8 ## 1.7552 1.1334 0.4429 0.3698 0.2454 0.1961 0.1751 0.1284boxplot(mod)Test for dispersions
set.seed(25)permutest(mod)## ## Permutation test for homogeneity of multivariate dispersions## Permutation: free## Number of permutations: 999## ## Response: Distances## Df Sum Sq Mean Sq F N.Perm Pr(>F) ## Groups 1 0.07931 0.079306 4.6156 999 0.045 *## Residuals 22 0.37801 0.017182 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1plot(mod)Test for dispersions
set.seed(4)permutest(mod, pairwise = TRUE)## ## Permutation test for homogeneity of multivariate dispersions## Permutation: free## Number of permutations: 999## ## Response: Distances## Df Sum Sq Mean Sq F N.Perm Pr(>F) ## Groups 1 0.07931 0.079306 4.6156 999 0.036 *## Residuals 22 0.37801 0.017182 ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## ## Pairwise comparisons:## (Observed p-value below diagonal, permuted p-value above diagonal)## grazed ungrazed## grazed 0.043## ungrazed 0.04295Test for locations with non-equal dispersion?
Marti Anderson & colleagues (2017) have proposed a solution that is related to the Berens-Fisher problem
This is in Primer but not yet in vegan
https://github.com/vegandevs/vegan/issues/344
Other stuff
Diagnostics for constrained ordinations
vegan provides a series of diagnostics to help assess the model fit
goodness()inertcomp()spenvcor()intersetcor()vif.caa()
Diagnostics | goodness of fit
goodness() computes a goodness of fit statistic for species or sites, controlled by argument display
Gives the cumulative proportion of variance explained by each axis
head(goodness(mods))## CCA1 CCA2 CCA3## Callvulg 0.0062471656 0.318907619 0.8254657## Empenigr 0.1164701677 0.137604904 0.1953245## Rhodtome 0.0999089739 0.169697909 0.1824153## Vaccmyrt 0.2361482843 0.240516323 0.2406730## Vaccviti 0.1523704591 0.156502301 0.2110550## Pinusylv 0.0009244423 0.004802076 0.0060096Diagnostics | inertia decomposition
inertcomp() decomposes the variance in samples or species in partial, constrained, and unconstrained components
statistic = "explained(default) gives the decomposition in terms of variancestatistic = "distance"gives decomposition in terms of the the residual distance
head(inertcomp(mods, proportional = TRUE))## CCA CA## Callvulg 0.8254657 0.1745343## Empenigr 0.1953245 0.8046755## Rhodtome 0.1824153 0.8175847## Vaccmyrt 0.2406730 0.7593270## Vaccviti 0.2110550 0.7889450## Pinusylv 0.0060096 0.9939904Diagnostics | species-environment correlations
spenvcor() returns the (weighted) correlation between the weighted average-based and the linear combination-based sets of site scores
A poor measure of goodness of fit. Sensitive to
- outliers (like all correlations)
- overfitting (using too many constraints)
Better models can have poorer species-environment correlations
spenvcor(mods)## CCA1 CCA2 CCA3 ## 0.8554793 0.8131627 0.8792221Diagnostics | interset correlations
intersetcor() returns the (weighted) correlation between the weighted average-based site scores and each constraint variable
Another poor diagnostic
- correlation based
- focuses on a single constraint--axis combination at a time
intersetcor(mods)## CCA1 CCA2 CCA3## Al 0.7356445 -0.1304293 0.4260453## P -0.3588931 -0.6109601 0.4478786## K -0.3767902 -0.1339051 0.7759566Vector fitting (envfit()) or biplot scores (scores(model, display = "bp")) are better alternatives
Links
I have several vegan-related posts on my blog.
For a list of posts see http://www.fromthebottomoftheheap.net/blog/
Acknowledgments
Funding
Fellow vegan maintainers
Jari Oksanen
Péter Sólymos
Slides
- HTML Slide deck bit.ly/adv-vegan © Simpson (2020)

- RMarkdown Source
References
- Anderson, M.J., 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46
- Anderson, M.J., 2006. Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62, 245–253
- Anderson, M.J., Walsh, D.C.I., 2013. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: What null hypothesis are you testing? Ecol. Monogr. 83, 557–574
- Anderson, M.J., Walsh, D.C.I., Robert Clarke, K., Gorley, R.N., Guerra-Castro, E., 2017. Some solutions to the multivariate Behrens-Fisher problem for dissimilarity-based analyses. Aust. N. Z. J. Stat. 59, 57–79
- Blanchet, F.G., Legendre, P., Borcard, D., 2008. Forward selection of explanatory variables. Ecology 89, 2623–2632
- Legendre, P., Anderson, M.J., 1999. Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecol. Monogr. 69, 1–24
- McArdle, B.H., Anderson, M.J., 2001. Fitting Multivariate Models to Community Data: A Comment on Distance-Based Redundancy Analysis. Ecology 82, 290–297
- Warton, D.I., Wright, S.T., Wang, Y., 2012. Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol. Evol. 3, 89–101